News
Navy cluster virus not related to strain doing the rounds in SL at that time

The KDU team at work in the laboratory
A surprise awaited KDU research team as they analysed raw data of samples – 9 were of a strain circulating in the US and the other from the B.1 lineage with mutations
Some interesting research with regard to the new coronavirus (SARS-CoV-2) strains found in Sri Lanka is emerging from the Faculty of Medicine of the General Sir John Kotelawala Defence University (KDU) at Ratmalana.
While one study focuses on a cluster which piqued the curiosity of all Sri Lankans in the early stage of the pandemic when the country had few cases last year, another follows a family’s journey during the Aluth Avurudda in April this year, before they fell like dominoes in the pathway of the virus.
It was around the time that the navy cluster erupted that the KDU RT-PCR testing laboratory saw the light of day initially in the Biomedical Lab-2 (BML-2) at the Faculty of Medicine, later moving to the University Hospital-KDU. The very first set of 58 samples from the navy cluster arrived at the lab on April 23, 2020 and to the team’s surprise, 26 samples were positive. This had been confirmed by independent testing at the Medical Research Institute (MRI) later.
Both the navy and family studies were led by Dr. Dharshan De Silva. The team which handled the navy study comprised Dr. Himali S. Jayasinghe arachchi, Dr. Prasad Premaratne, Prof. Charitha Goonasekera, Consultant Microbiologist Dr. Dilini Nakawita and Consultant Physician Dr. Dumitha Govindapala who were supported by the technical staff: Teshan Chathuranga, Pawani Kawyangana and Upeksha Kulasekara.

Dr. Dharshan De Silva
This was while the in-depth genetic analysis of virus sequences was carried out by Dr. Jayasinghearachchi, with the Next Generation Sequencing (NGS) being handled by Genelabs Medical (Pvt.) Ltd., Narahenpita, which provided the raw data to the KDU team. The research work including sequencing of the stored samples from the navy cluster was carried out this year (2021) after securing a KDU grant.
Since the pandemic took hold of the world, the first imported case had been detected in Sri Lanka on January 29 and the first local infection on March 11. The number of positive cases was 199 up to late April 2020, until the navy cluster mushroomed.
As Sri Lanka wondered how day by day the navy cluster ballooned, finally becoming a ‘big’ 950 infections, Dr. De Silva says they couldn’t believe their eyes when the 26 samples were positive. It was then that the team decided to follow-up with genetic sequencing and raw data analysis of 10 samples later and for this purpose stored these samples.
This genome analysis included bioinformatics, mutation profiling and phylogenetics (the study of evolutionary relationships) to establish the evolutionary pattern of the virus.
“We compared the data from the navy with those available not only in Sri Lanka but also from other countries,” says Dr. Jayasinghearachchi, explaining that they wanted to identify the closest relatives of what was unearthed in the navy cluster.
Giving the backdrop, she says that it was the time of the first wave and the predominant variant of the new coronavirus across the world was D614G. It had started with 13% of infections in China alone and then taken control of 70% of the infections globally.
The team’s findings were “very” interesting. The 10 samples of the navy cluster did not show any connection to the existing strain in Sri Lanka at that time.

Prof. Charitha Goonasekera
Nine samples were of a strain of the B.1.3 lineage, which was circulating mostly in the United States of America during the early part of the pandemic.
Variants come about when there are mutations in the viral RNA (ribonucleic acid), with mutations of significance mainly occurring in the spike protein.
Dr. Jayasinghearachchi details what they found:
- The D614G mutation with the spike protein amino acid change. This was and even currently is a ‘dominant’ mutation which increases infectivity.Three prominent ‘linkage mutations’ co-occurring with the D614G mutation – the P323L mutation in the viral polymerase; the Q57H changes in the ORF3a region of the viral genome; and the T85I mutation in the non-structural protein 2.

Dr. Himali S. Jayasinghe- arachchi
The 10th sample, meanwhile, had evolved from the parent lineage of B.1 but also had the same D614G mutation and the three ‘linkage mutations’ along with other common mutations.
She says that the navy cluster may have been set off by an importation of this variant, through travellers coming to Sri Lanka but it is usually very difficult to establish the origins.

Dr. Dilini Nakawita
Compared to the other strains circulating in Sri Lanka at that time, the transmissibility and infectivity of these strains were high but due to the lockdown and prompt action taken by the navy, the strain did not spread beyond this cohort.
The navy cluster analysis may be revealing an old tale, but the study team is hopeful that the establishment of these facts would be of interest to researchers peering closely at the evolution of the new coronavirus.
Immunology studies on the 26 samples of the navy cohort are ongoing.
(Next: The family study & patients’ core infections) Box within this copy
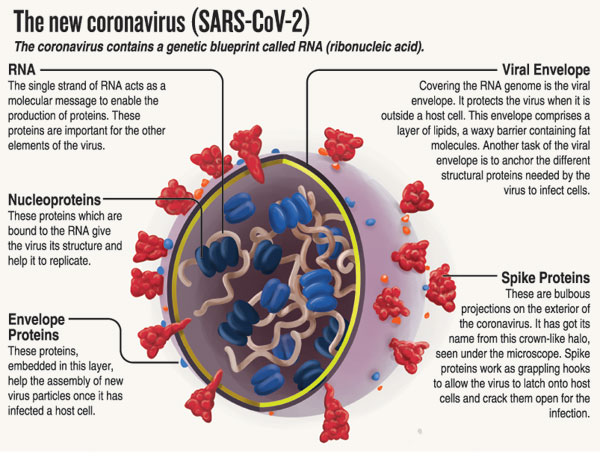
| Mutations and variants All viruses – including SARS-CoV-2, the virus that causes COVID-19 – evolve over time. When a virus replicates or makes copies of itself, it sometimes changes a little bit, which is normal for a virus. These changes are called “mutations”. A virus with one or more new mutations is referred to as a “variant” of the original virus, according to the World Health Organization. It adds: “When a virus is widely circulating in a population and causing many infections, the likelihood of the virus mutating increases. The more opportunities a virus has to spread, the more it replicates – and the more opportunities it has to undergo changes. “Most viral mutations have little to no impact on the virus’s ability to cause infections and disease. But depending on where the changes are located in the virus’s genetic material, they may affect a virus’s properties, such as transmission (it may spread more or less easily) or severity (it may cause more or less severe disease).” Meanwhile, the four dominant variants of SARS-CoV-2 spreading among global populations currently are:
| |

